New findings in cardiomyopathy
Cardiomyopathy is a relatively rare and refractory myocardial disease caused by genetic and environmental factors. Cardiomyopathy is classified into dilated cardiomyopathy, hypertrophic cardiomyopathy, arrhythmogenic right ventricular cardiomyopathy, restrictive cardiomyopathy, and unclassified cardiomyopathy. Major risk factors includes family history of cardiomyopathy, coronary heart disease, obesity, long term high blood pressure, alcoholism, chronic kidney disease and diabetes. Clarification of the genotype–phenotype correlation is needed to develop the precision medicine in cardiomyopathy. In addition, elucidation of the mechanisms involved in cardiomyopathy onset and progression from the causative genes will lead to the development of novel therapeutic agents.
1. Cardiomyocyte-specific knockout of ADAM17 ameliorates left ventricular remodeling and function in diabetic cardiomyopathy
A subset of diabetic patients exhibited cardiac systolic and diastolic dysfunction without hypertension and coronary artery disease, labeled diabetic cardiomyopathy (DCM). Angiotensin-converting enzyme 2 (ACE2) has proven beneficial in attenuating diabetic cardiomyopathy (DCM) but has been found to be a substrate of a disintegrin and metalloprotease protein-17 (ADAM17). Cheng Zhang, Department of Cardiology, Qilu Hospital, Cheeloo College of Medicine, Shandong University, China, and his team used differentiated H9c2 cells and neonatal rat cardiomyocytes (NRCMs) to explore the molecular mechanisms underlying the effect of ADAM17 on DCM[1]. Cardiomyocyte-specific knockout of ADAM17 mitigated cardiac fibrosis and cardiomyocyte apoptosis and ameliorated cardiac dysfunction in mice with DCM. Bioinformatic analyses detected a number of genes enriched in metabolic pathways, in particular the AMPK signaling pathway, expressed differentially between the hearts of cardiomyocyte-specific knockout of ADAM17 diabetic mice. The mechanism may involve activated AMPK pathway, increased autophagosome formation and improved autophagic flux (Fig.1), which reduced the apoptotic response in cardiomyocytes. Thus, inhibition of ADAM17 may provide a promising approach to the treatment of DCM.
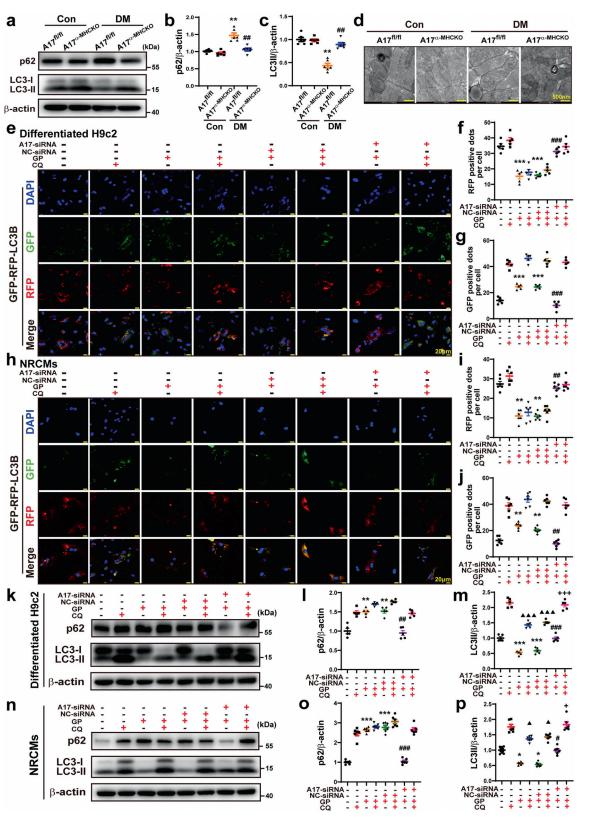
Fig.1 Effects of ADAM17 deficiency on cardiomyocyte autophagy in vivo and in vitro and on autophagic flux in differentiated H9c2 cells and NRCMs
2. Pathogenic variants damage cell composition and single-cell transcription in cardiomyopathies
Pathogenic variants in genes that cause dilated cardiomyopathy (DCM) and arrhythmogenic cardiomyopathy (ACM) convey high risks for the development of heart failure through unknown mechanisms. Using single-nucleus RNA sequencing, CHRISTINE E. SEIDMAN, Department of Genetics, Harvard Medical School, Boston, USA, and his team characterized the transcriptome of 880,000 nuclei from 18 control and 61 failing, nonischemic human hearts with pathogenic variants in DCM and ACM genes or idiopathic disease[2]. They identified 10 major cell types and 71 distinct transcriptional states. DCM and ACM tissues showed significant depletion of cardiomyocytes and increased endothelial and immune cells. Fibrosis was expanded in disease hearts, but, unexpectedly, fibroblasts were not increased, and instead showed altered transcriptional states that indicated activated remodeling of the extracellular matrix. Through analyses of receptor and ligand expression across all cells, they observed changes in intercellular signaling and communications, such as increased endothelin signaling in LMNA hearts, tumor necrosis factor in PKP2 hearts, and others(Fig.2). These data provide candidate therapeutic targets for future research and interventional opportunities to improve and personalize treatments for cardiomyopathies and heart failure.
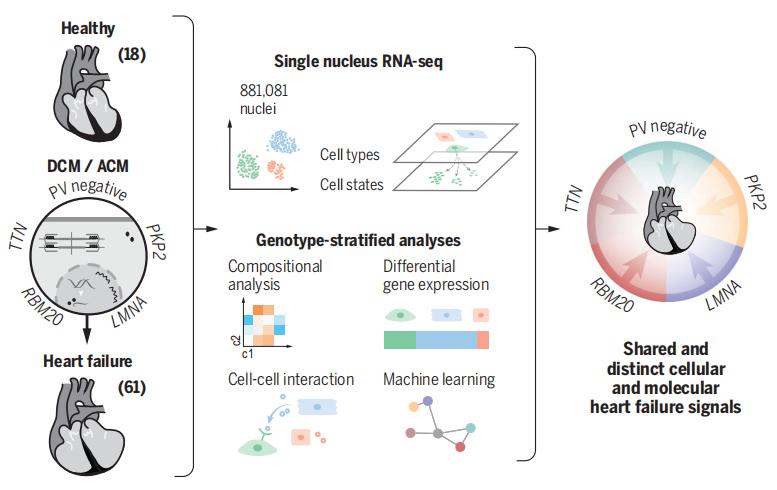
Fig.2 Genotype-stratified analyses of heart failure at the single-nuclei level
3. Single-nucleus profiling of human dilated and hypertrophic cardiomyopathy
Patrick T. Ellinor, Cardiovascular Research Center, Massachusetts General Hospital, USA, and his team identified extensive molecular alterations in failing hearts at single-cell resolution by performing single-nucleus RNA sequencing of nearly 600,000 nuclei in left ventricle samples from 11 hearts with dilated cardiomyopathy and 15 hearts with hypertrophic cardiomyopathy as well as 16 non-failing hearts[3]. The transcriptional profiles of dilated or hypertrophic cardiomyopathy hearts broadly converged at the tissue and cell-type level. Further, a subset of hearts from patients with cardiomyopathy harbour a unique population of activated fibroblasts that is almost entirely absent from non-failing samples. They performed a CRISPR-knockout screen in primary human cardiac fibroblasts to evaluate this fibrotic cell state transition; knockout of genes associated with fibroblast transition resulted in a reduction of myofibroblast cell-state transition upon TGFβ1 stimulation for a subset of genes(Fig.3). The results provide insights into the transcriptional diversity of the human heart in health and disease as well as new potential therapeutic targets and biomarkers for heart failure.
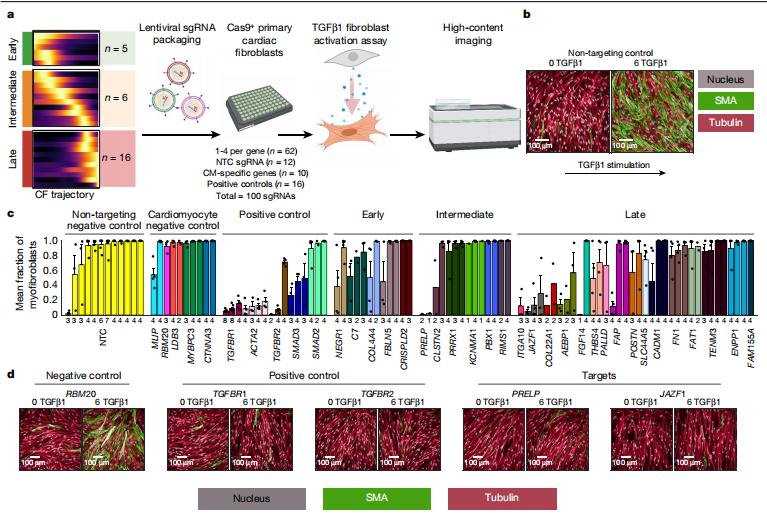
Fig.3 Cellular assay of myofibroblast transition in cardiac fibroblasts
4. SIRPα Mediates IGF1 Receptor in Cardiomyopathy Induced by Chronic Kidney Disease
Chronic kidney disease (CKD) is characterized by increased myocardial mass despite near-normal blood pressure, suggesting the presence of a separate trigger. Sandhya S. Thomas, Nephrology Division, Department of Medicine, Baylor College of Medicine, USA, and his team assessed the role of circulating SIRPα in CKD-induced adverse cardiac remodeling[4]. They demonstrated that SIRPα regulates myocardial insulin/IGF1R (insulin growth factor-1 receptor) signaling in CKD. Both muscle-specific or cardiac muscle–specific SIRPα KO mice did not significantly activate fetal genes and maintained insulin/IGF1R signaling with suppressed fibrosis despite the presence of CKD(Fig.4). SIRPα directly interacted with IGF1R. Next, rSIRPα (recombinant SIRPα) protein was introduced into muscle-specific SIRPα KO mice reestablishing the insulin/IGF1R signaling activity. Additionally, overexpression of SIRPα in myoblasts and cardiomyocytes impaired pAKT (phosphorylation of AKT) and insulin/IGF1R signaling. Their discovery suggests novel targets for therapies to potentially prevent CKD-associated heart failure.
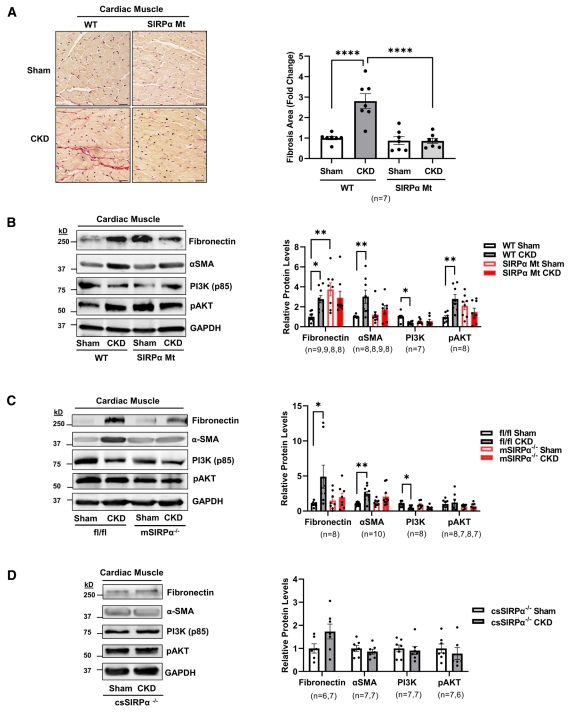
Fig.4 Suppressing SIRPα (signal regulatory protein alpha) prevents cardiac fibrosis
References
[1]Xue F, Cheng J, Liu Y, et al. Cardiomyocyte-specific knockout of ADAM17 ameliorates left ventricular remodeling and function in diabetic cardiomyopathy of mice [J]. Signal Transduct Target Ther. 2022, 7(1):259. (IF=38.104)
[2]Reichart D, Lindberg EL, Maatz H, et al. Pathogenic variants damage cell composition and single cell transcription in cardiomyopathies [J]. Science. 2022, 377(6606):eabo1984. (IF=63.714)
[3]Chaffin M, Papangeli I, Simonson B, et al. Single-nucleus profiling of human dilated and hypertrophic cardiomyopathy [J]. Nature. 2022, 608(7921):174-180. (IF=69.504)
[4]Thomas SS, Wu J, Davogustto G, et al. SIRPα Mediates IGF1 Receptor in Cardiomyopathy Induced by Chronic Kidney Disease [J]. Circ Res. 2022, 131(3):207-221. (IF=23.213)
Cloud-Clone can not only provide a variety of cardiovascular system disease models, including cardiac hypertrophy, myocardial infarction, arrhythmia, myocarditis, heart failure, etc., covering common cardiovascular system diseases. We also have a variety of common indicators of cardiovascular system signaling pathway and the above mentioned ADAM17, AMPK, LMNA, PKP2, SIRPα, IGF1R and other related products, which can help the majority of scientific researchers to study cardiovascular system diseases.


