New findings on the mechanism of progression and metastasis of colorectal cancer
Colorectal cancer (CRC) is one of the most common human malignancies accounting for approximately 10% of worldwide cancer incidence and mortality. While early-stage CRC is curable by surgery, treatment of metastatic colorectal cancer (mCRC) remains an unmet clinical need. Moreover, about 25% of CRC cases are diagnosed only at the metastatic stage. Despite the extensive molecular and functional knowledge on this disease, systemic therapy for mCRC still relies on traditional 5-fuorouracil (5-FU)-based chemotherapy regimens. So far, targeted therapies and immunotherapy have shown effectiveness only in a limited subset of CRC patients. Therefore, there is an urgent need to understand the molecular mechanisms and regulations underlying CRC development and treatment resistance in order to implement novel, rationally driven, tailored therapies.
1. AP4 suppresses DNA damage and contributes to 5-FU resistance via inducing MDC1 and repressing miR-22-3p
AP4 encodes a basic helix-loop-helix leucine zipper (bHLH-LZ) transcription factor and is a direct target gene of the oncogenic transcription factor c-MYC. Heiko Hermeking, Experimental and Molecular Pathology, Institute of Pathology, LudwigMaximilians-University, Germany, and his team set out to determine the relevance of AP4 in human CRC cells[1]. A CRISPR/Cas9 approach was employed to generate AP4-defcient CRC cell lines with inducible expression of c-MYC. Inactivation of AP4 in CRC cell lines resulted in increased spontaneous and c-MYC-induced DNA damage, chromosomal instability (CIN) and cellular senescence(Fig.1). AP4-defcient cells displayed increased expression of the long non-coding RNA MIR22HG, which encodes miR-22-3p and was directly repressed by AP4. Furthermore, Mediator of DNA damage Checkpoint 1 (MDC1), a central component of the DNA damage response and a known target of miR-22-3p, displayed decreased expression in AP4-defcient cells. Inhibition of miR-22-3p or ectopic MDC1 expression reversed the increased senescence, DNA damage, CIN and defective HR observed in AP4-defcient CRC cells. AP4-defciency also sensitized CRC cells to 5-FU treatment, whereas ectopic AP4 conferred resistance to 5-FU in a miR-22-3p and MDC1-dependent manner. These fndings explain how elevated AP4 expression contributes to development and chemo-resistance of colorectal cancer after c-MYC activation.
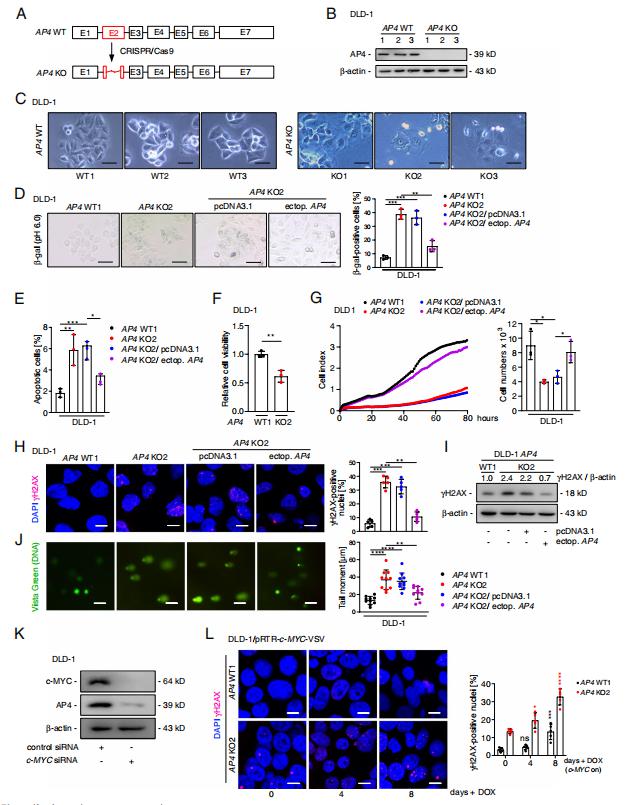
Fig.1 AP4 inactivation induces DNA damage and senescence in CRC cells
2. Interaction of lncRNA MIR100HG with hnRNPA2B1 facilitates m6A-dependent stabilization of TCF7L2 mRNA and CRC progression
Epithelial-to-mesenchymal transition (EMT) is a process linked to metastasis and drug resistance with non-coding RNAs (ncRNAs) playing pivotal roles. Yuanyuan Lu, State Key Laboratory of Cancer Biology, National Clinical Research Center for Digestive Diseases, Xijing Hospital of Digestive Diseases, Fourth Military Medical University, China, and his team investigated the role of MIR100HG in CRC cetuximab resistance or EMT[2]. The expression of MIR100HG was strongly correlated with EMT markers and acted as a positive regulator of EMT. MIR100HG sustained cetuximab resistance and facilitated invasion and metastasis in CRC cells both in vitro and in vivo. hnRNPA2B1 was identified as a binding partner of MIR100HG. Mechanistically, MIR100HG maintained mRNA stability of TCF7L2, a major transcriptional coactivator of the Wnt/β-catenin signaling, by interacting with hnRNPA2B1(Fig.2). hnRNPA2B1 recognized the N6-methyladenosine (m6A) site of TCF7L2 mRNA in the presence of MIR100HG. The MIR100HG/hnRNPA2B1/TCF7L2 axis was augmented in specimens from CRC patients who either developed local or distant metastasis or had disease progression that was associated with cetuximab resistance. The findings identified MIR100HG as a potent EMT inducer in CRC that may contribute to cetuximab resistance and metastasis by activation of a MIR100HG/hnRNPA2B1/TCF7L2 feedback loop.
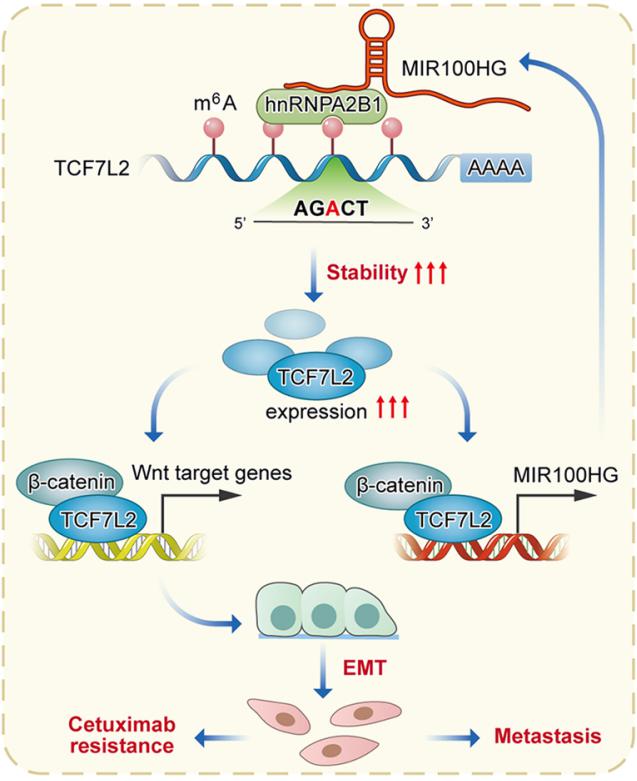
Fig.2 Interaction of lncRNA MIR100HG with hnRNPA2B1 facilitates m6A-dependent stabilization of TCF7L2 mRNA and CRC progression
3. PIK3CA mutations-mediated downregulation of circLHFPL2 inhibits CRC progression via upregulating PTEN
PIK3CA mutation and PTEN suppression lead to tumorigenesis and drug resistance in CRC. Yong Gao, Department of Oncology, Shanghai East Hospital, Tongji University School of Medicine, China, and his team investigated the role of circLHFPL2 in regulating PIK3CA mutations and MEK inhibitor resistance in CRC[3]. RNA sequencing defined downregulated expression of circLHFPL2 in both PIK3CAH1047R (HCT116) and PIK3CAE545K (DLD1) cells. CircLHFPL2 was also downregulated in PIK3CA-mutant CRC primary cells and tissues, which was correlated with poor prognosis. CircLHFPL2 was mainly localized in the cytoplasm and its downregulation was attributed to the PI3K/AKT signaling pathway activated by phosphorylating Foxo3a. CircLHFPL2 inhibited PI3KCA-Mut CRC progression both in vitro and in vivo. Furthermore, their work indicated that circLHFPL2 acts as a ceRNA to sponge miR-556-5p and miR-1322 in CRC cells and in turn modulate the expression of PTEN(Fig.3). In addition, downregulation of circLHFPL2 leads to MEK inhibitor resistance in CRC. Therefore, targeting circLHFPL2 could be an efective approach for the treatment of CRC patients harboring oncogenic PIK3CA mutations.
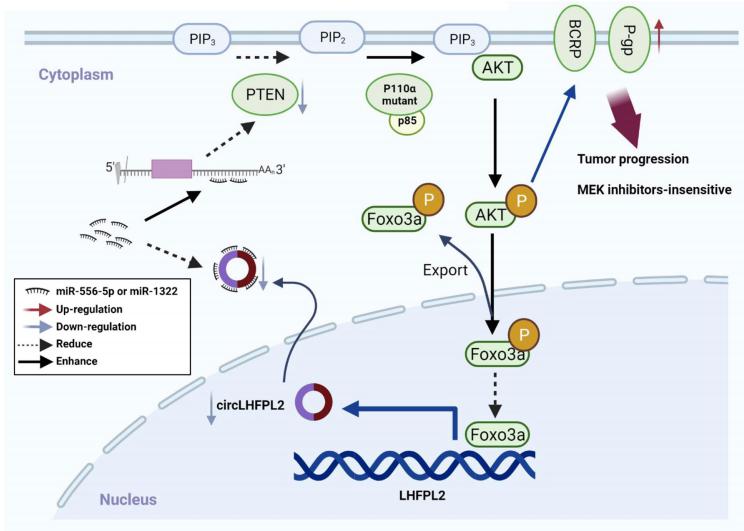
Fig.3 Schema of the positive feedback loop of sustained activation of PI3K/AKT via circLHFPL2/miR-556-5p and miR-1322/PTEN axis and downregulation of circLHFPL2 enhanced MEK inhibitor resistance by upregulating P-gp and BCRP in PIK3CA-mutant CRC cells
4. The m6A modifcation of circALG1 promotes the metastasis of colorectal cancer mediated by the miR-342-5p/PGF signalling pathway
Gui Hu, Department of Gastrointestinal Surgery, The Third Xiangya Hospital of Central South University, China, and his team used a human circRNA microarray to detect the expression profles of circRNAs in cancer and paracancerous tissues from patients with colorectal cancer (CRC) and peripheral blood specimens from patients with CRC and healthy individuals[4]. CircALG1 was highly expressed in both the peripheral blood and tumour tissues of patients with CRC and was closely associated with CRC metastasis. CircALG1 overexpression promoted the migration and invasion of CRC cells, and circALG1 silencing and reduction of the circALG1 m6A modifcation level inhibited CRC cell migration and invasion. In vivo experiments further confrmed the prometastatic role of circALG1 in CRC. Further mechanistic studies showed that circALG1 upregulated the expression of placental growth factor (PGF) by binding to miR-342-5p and that m6A modifcation enhanced the binding of circALG1 to miR-342-5p and promoted its ceRNA function(Fig.4). These results suggested that circALG1 could be a potential therapeutic target in and a prognostic marker for CRC.
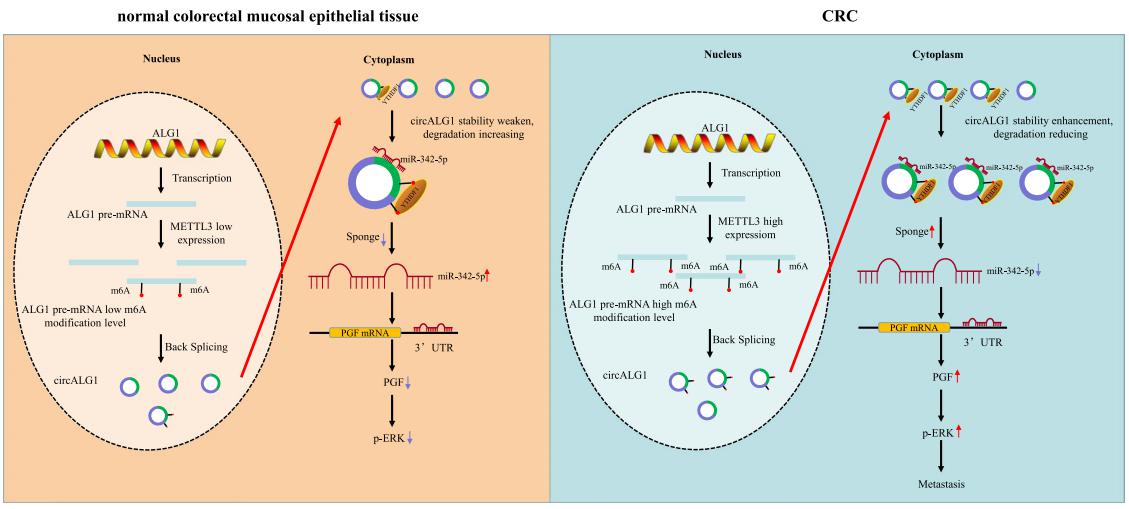
Fig.4 CircALG1 relieves the inhibitory efect of miR-342-5p on PGF mRNA expression by competitively binding to miR-342-5p and promotes PGF expression to result in enhanced invasion of CRC
References
[1]Chou J, Kaller M, Jaeckel S, Rokavec M, Hermeking H. AP4 suppresses DNA damage, chromosomal instability and senescence via inducing MDC1/Mediator of DNA damage Checkpoint 1 and repressing MIR22HG/miR-22-3p[J]. Mol Cancer. 2022,21(1):120. (IF=27.401)
[2]Liu H, Li D, Sun L, et al. Interaction of lncRNA MIR100HG with hnRNPA2B1 facilitates m6A-dependent stabilization of TCF7L2 mRNA and colorectal cancer progression[J]. Mol Cancer. 2022,21(1):74. (IF=27.401)
[3]Chong X, Chen J, Zheng N, et al. PIK3CA mutations-mediated downregulation of circLHFPL2 inhibits colorectal cancer progression via upregulating PTEN[J]. Mol Cancer. 2022,21(1):118. (IF=27.401)
[4]Lin C, Ma M, Zhang Y, et al. The N6-methyladenosine modification of circALG1 promotes the metastasis of colorectal cancer mediated by the miR-342-5p/PGF signalling pathway [J]. Mol Cancer. 2022,21(1):80. (IF=27.401)
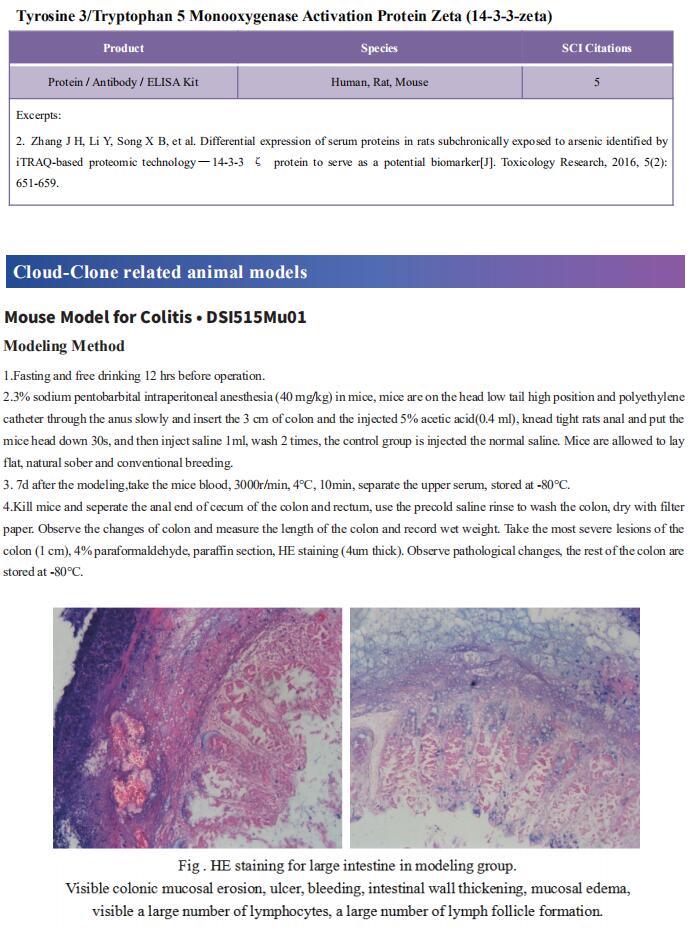
Cloud-Clone can not only provide a variety of experimental tumor animal models, including tumor transplantation animal model, spontaneous tumor animal model, induced tumor animal model, tumor metastasis animal model, covering common tumor research. We also have various cancer detection indicators and the above-mentioned Wnt/β-catenin, PI3K/AKT pathway related products, which can help the general scientific researchers to carry out cancer related research.