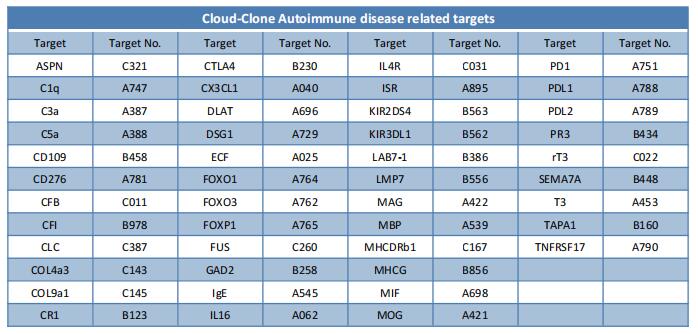
New discovery in genetic study of systemic lupus erythematosus
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease, which damages multiple tissues and organs, resulting from the production of autoantibodies to nuclear antigens. The clinical manifestations of SLE are highly heterogeneous, including cutaneous, musculoskeletal, renal, hematologic, neurologic, and other diverse symptoms. To date, nearly 180 genomic loci have been identifed as associated with SLE susceptibility in genetic studies in multiple ancestries. Recent genetic studies leverage diverse cell type–specifc epigenetic resources and other biological resources to draw better pictures of the disease pathogenesis, thus updating the genetic architecture of SLE. Such genetic fndings can be used to estimate a degree of genetic risk for SLE and to provide more efective drug targets.
1. Genomic sequencing and functional analyses identify MAP4K3/GLK germline and somatic variants associated with SLE
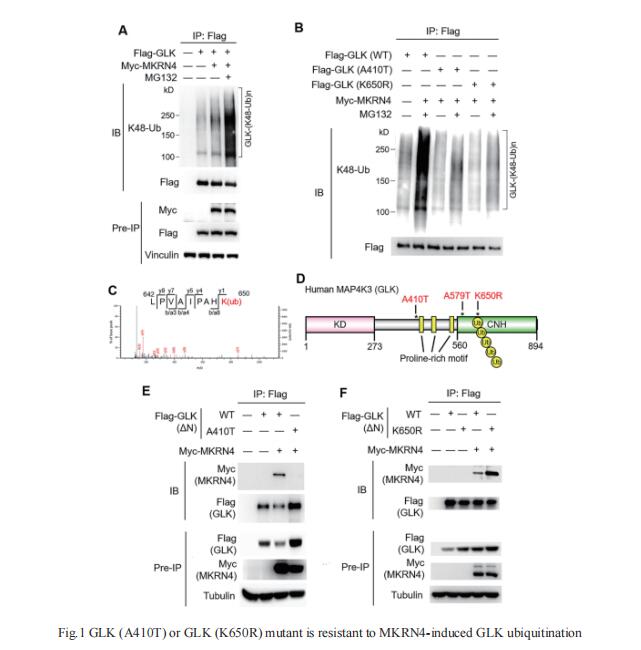
MAP4K3 (GLK) overexpression in T cells induces interleukin (IL)-17A production and autoimmune responses. GLK overexpressing T-cell population is correlated with severity of human SLE; however, it is unclear how GLK is upregulated in patients with SLE. Tse-Hua Tan, Immunology Research Center, National Health Research Institutes, Taiwan, and his team enrolled 181 patients with SLE and 250 individuals without SLE in two independent cohorts from different hospitals/cities[1]. Genomic DNAs of peripheral blood mononuclear cells were subjected to next-generation sequencing to identify GLK gene variants. They identified 58 patients with SLE from Cohort #1 and #2 with higher frequencies of a somatic variant in GLK 3′-UTR; these patients with SLE showed increased serum anti-double-stranded DNA levels and decreased serum C3/C4 levels. This somatic variant in 3′-UTR enhanced GLK mRNA levels in T cells. In addition, they identified five patients with SLE with GLK (A410T) germline variant in Cohort #1 and #2, as well as two other patients with SLE with GLK (K650R) germline variant in Cohort #1. Both GLK (A410T) and GLK (K650R) mutants inhibited GLK ubiquitination induced by the novel E3 ligase makorin ring-finger protein 4 (MKRN4), leading to GLK protein stabilisation(Fig.1). Multiple GLK germline and somatic variants cause GLK induction by increasing mRNA or protein stability in patients with SLE. Taken together, individuals who harbour the aforementioned GLK germline or somatic variants may be at high risk for SLE.
2. Single-cell RNA-seq reveals cell type–specific molecular and genetic associations to lupus
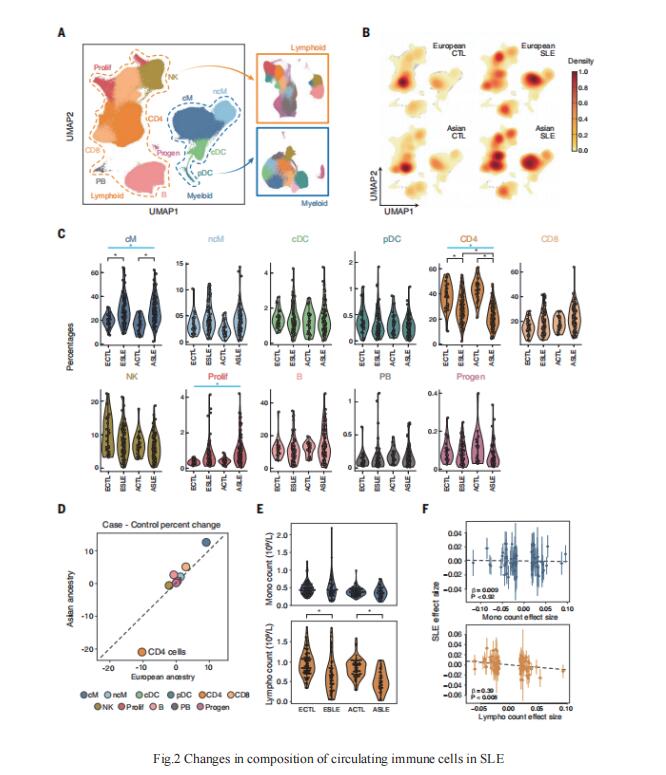
Despite the progress in the study of SLE related loci, a comprehensive census of circulating immune cells in SLE remains incomplete, and annotating the cell types and contexts that mediate genetic associations remains challenging. Chun Jimmie Ye, Division of Rheumatology, Department of Medicine, University of California, USA, and his team used multiplexed single-cell RNA sequencing (mux-seq) to profile more than 1.2 million PBMCs from 162 SLE cases and 99 healthy controls of either Asian or European ancestry[2]. Cases exhibited elevated expression of type 1 interferon–stimulated genes (ISGs) in monocytes, reduction of naïve CD4+ T cells that correlated with monocyte ISG expression, and expansion of repertoire-restricted cytotoxic GZMH+ CD8+ T cells(Fig.2). They integrated dense genotyping data to map cell type– specific cis–expression quantitative trait loci and to link SLE-associated variants to cell type–specific expression. These results demonstrate mux-seq as a systematic approach to characterize cellular composition, identify transcriptional signatures, and annotate genetic variants associated with SLE.
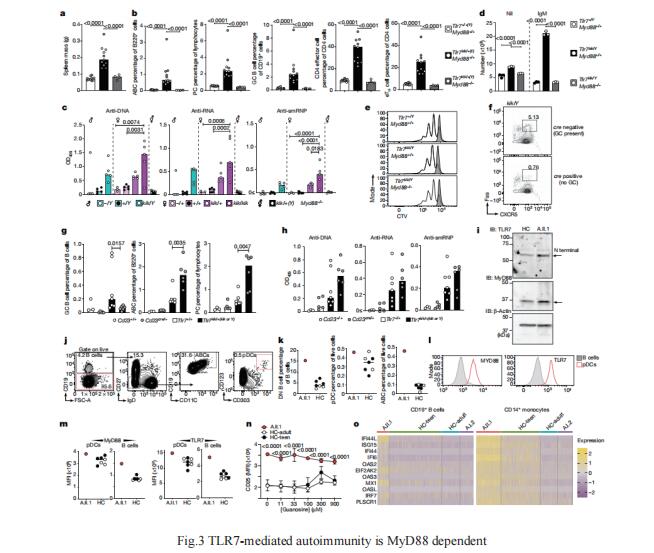
3. TLR7 gain-of-function genetic variation causes human lupus
Although circumstantial evidence supports enhanced Toll-like receptor 7 (TLR7) signalling as a mechanism of human systemic autoimmune disease, evidence of lupus-causing TLR7 gene variants is lacking. Carola G. Vinuesa, Centre for Personalised Immunology, Department of Immunology and Infectious Disease, John Curtin School of Medical Research, Australian National University, Australia, and his team describe human systemic lupus erythematosus caused by a TLR7 gain-of-function variant[3]. They identifed a de novo, previously undescribed missense TLR7Y264H variant in a child with severe lupus. The TLR7Y264H variant selectively increased sensing of guanosine and 2',3'-cGMP, and was sufcient to cause lupus when introduced into mice. They showed that enhanced TLR7 signalling drives aberrant survival of B cell receptor-activated B cells, and in a cell-intrinsic manner, accumulation of CD11c+ age-associated B cells and germinal centre B cells. Defciency of MyD88 (an adaptor protein downstream of TLR7) rescued autoimmunity, aberrant B cell survival, and all cellular and serological phenotypes(Fig.3). The study established the importance of TLR7 and guanosine-containing self-ligands for human lupus pathogenesis, which paves the way for therapeutic TLR7 or MyD88 inhibition.
References
[1]Chuang HC, Hung WT, Chen YM, et al. Genomic sequencing and functional analyses identify MAP4K3/GLK germline and somatic variants associated with systemic lupus erythematosus [J]. Ann Rheum Dis. 2022, 81(2):243-254. (IF=19.103)
[2]Perez RK, Gordon MG, Subramaniam M, et al. Single-cell RNA-seq reveals cell type-specific molecular and genetic associations to lupus [J]. Science. 2022, 376(6589):eabf1970. (IF=47.728)
[3]Brown GJ, Cañete PF, Wang H, et al. TLR7 gain-of-function genetic variation causes human lupus [J]. Nature. 2022, 605(7909):349-356. (IF=49.967)
Cloud-Clone can not only provide animal models of various autoimmune diseases, including systemic lupus erythematosus, rheumatoid arthritis, psoriasis, scleroderma, etc., covering common autoimmune diseases; we also have all kinds of autoimmune disease detection indicators, which can help the majority of scientific researchers to conduct autoimmune disease research.